Matsoukas M-T, et. al. Design of Small Non-Peptidic Ligands That Alter Heteromerization between Cannabinoid CB1 and Serotonin 5HT2A Receptors. Journal of Medicinal Chemistry. 2025, 68, 261.
Matsoukas M-T, et. al. Identification of Small-Molecule Antagonists Targeting the Growth Hormone Releasing Hormone Receptor (GHRHR). Journal of Chemical Information and Modeling. 2024, 64, 7056.
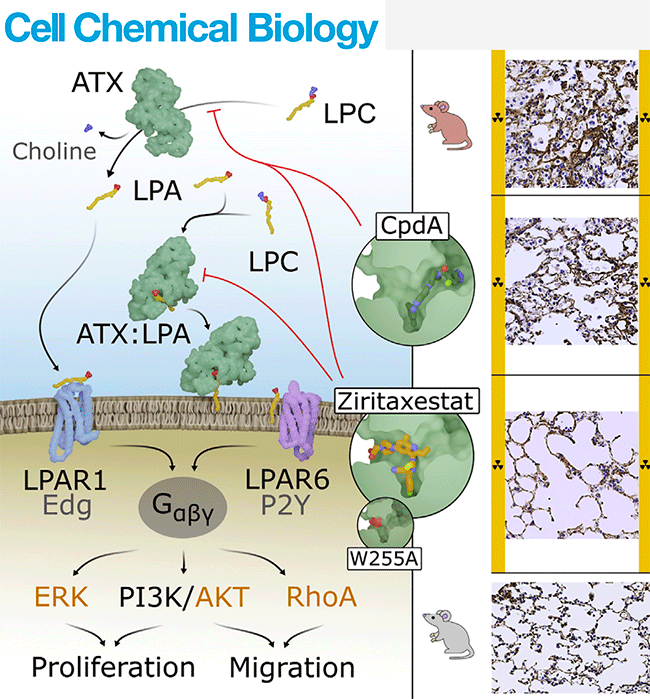
Salgado-Polo F, et. al., Autotaxin facilitates selective LPA receptor signaling. Cell Chemical Biology, 2023, 30, 69.
Sánchez-Morales A, et. al., Design and synthesis of a novel non peptide CN-NFATc signaling inhibitor for tumor suppression in triple negative breast cancer. European Journal of Medicinal Chemistry, 2022, 238, 114514.
Swarbrick C, et. al., Amidoxime prodrugs convert to potent cell-active multimodal inhibitors of the dengue virus protease. European Journal of Medicinal Chemistry, 2021, 224, 113695.
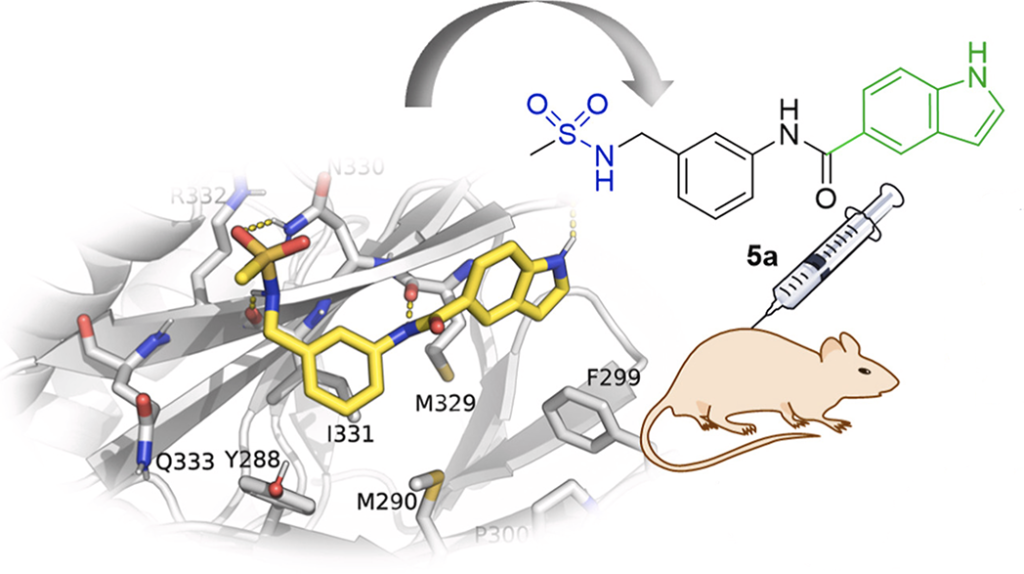
Busch J, et. al. Identification of compounds that bind the centriolar protein SAS-6 and inhibit its oligomerisation. Journal of Biological Chemistry. 2020, 295, 17922.
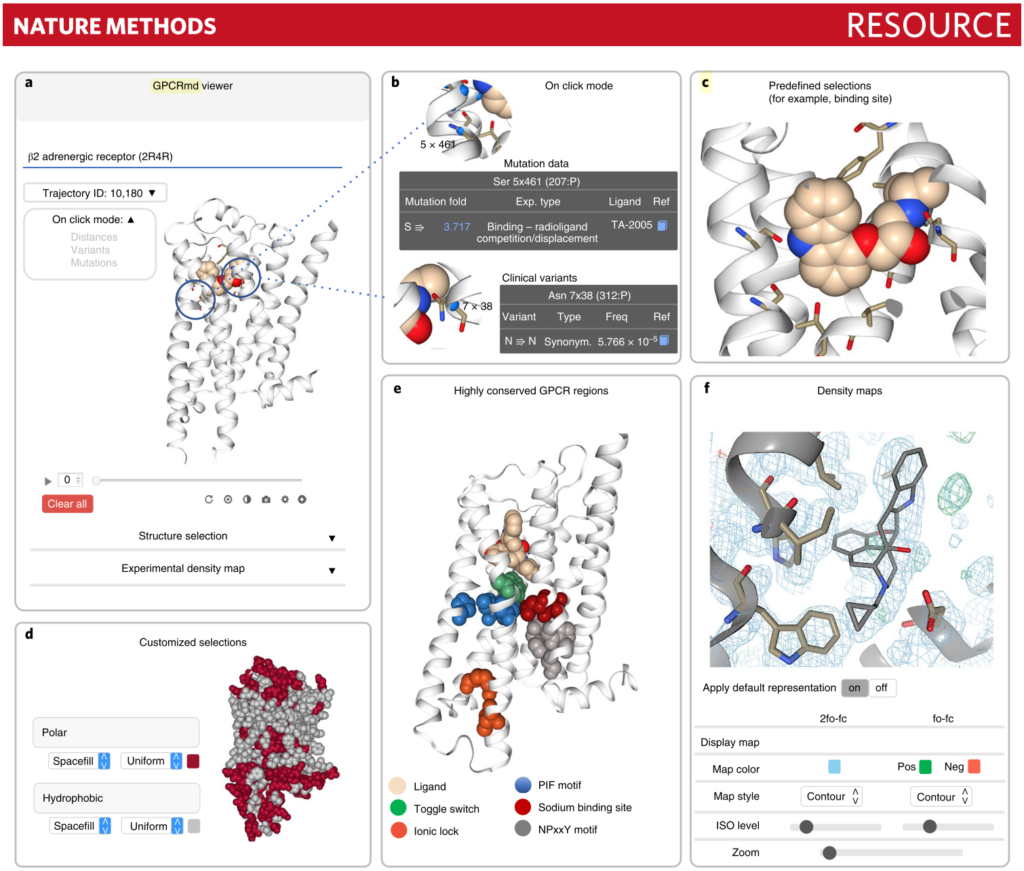
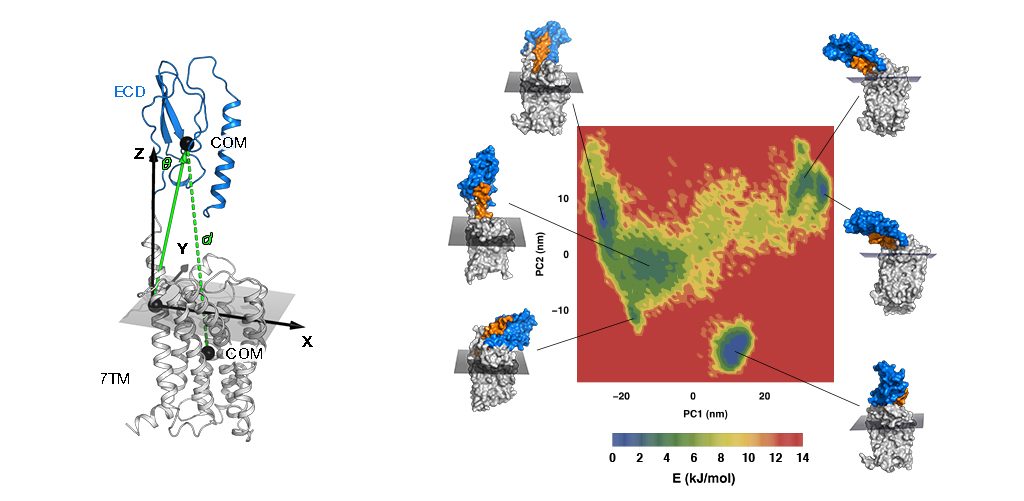
Rodríguez-Espigares I, et. al. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nature Methods. 2020, 17, 777.
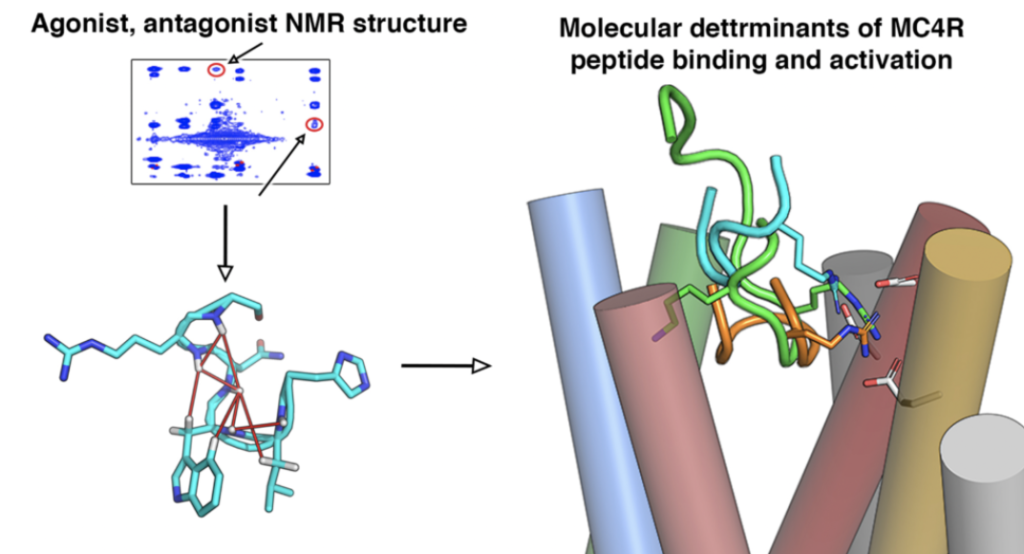
Zachmann J, et. al. A Combined Computational and Structural Approach into Understanding the Role of Peptide Binding and Activation of the Melanocortin Receptor 4, Journal of Chemical Information and Modeling. 2020, 60, 1461.
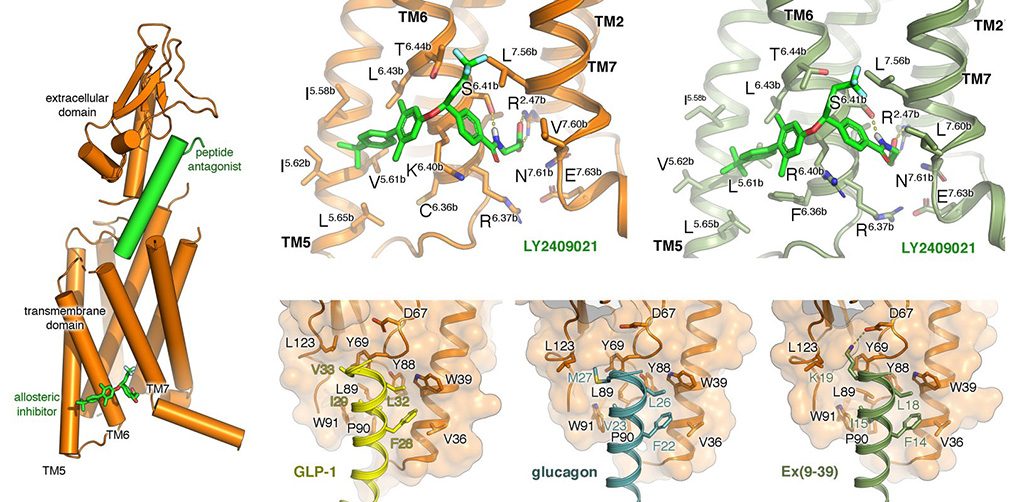
Chepurny OG‡, et. al. Non-conventional glucagon and GLP-1 receptor agonist and antagonist interplay at the GLP-1 receptor revealed in high-throughput FRET assays for cAMP. Journal of Biological Chemistry, 2019, (in press).
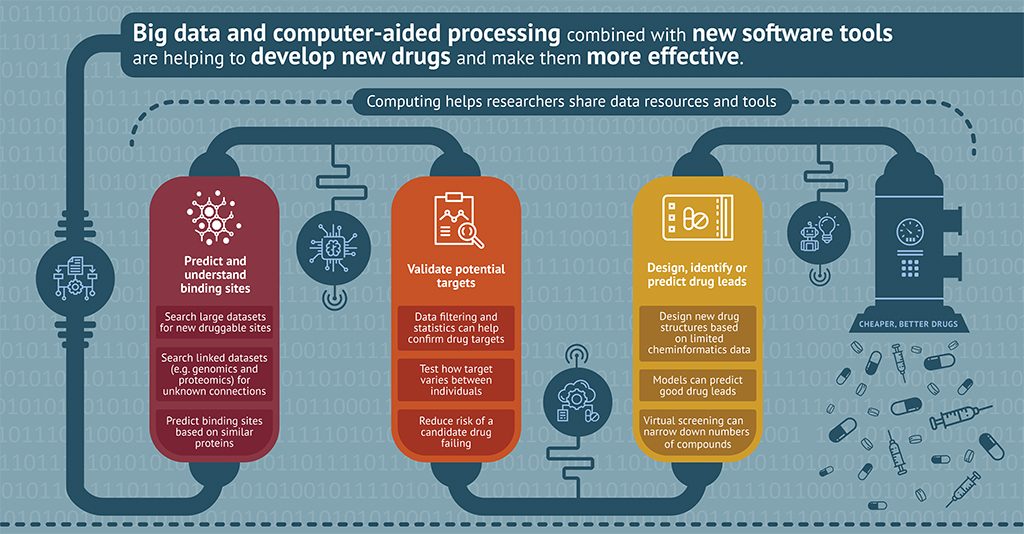
Katsila T, et. al., Ηοw far have we come with contextual data integration in drug discovery? Expert Opinion on Drug Discovery, 2018, 13, 791.
Salgado-Polo F, et. al., Lysophosphatidic acid produced by Autotaxin acts as an allosteric modulator of its catalytic efficiency. Journal of Biological Chemistry, 2018, 293, 14312.
Perez-Benito L, et. al., The Size Matters? A Computational Tool to Design Bivalent Ligands. Bioinformatics, 2018, bty422.
Matsoukas M-T, Spyroulias GA. Dynamic properties of the growth hormone releasing hormone receptor (GHRHR) and molecular determinants of GHRH binding. Molecular BioSystems, 2017, 13, 1313.
Katsila T, et. al., Computational approaches in target identification and drug discovery. Computational and Structural Biotechnology Journal, 2016, 14, 177.
Matsoukas M-T, et. al., Identification of small-molecule inhibitors of calcineurin-NFATc signaling that mimic the PxIxIT motif of calcineurin binding partners. Science Signaling, 2015, 8, 63.
Cordomí A, et. al., Functional elements of the gastric inhibitory polypeptide receptor: Comparison between secretin- and rhodopsin-like G protein-coupled receptors. Biochemical Pharmacology,2015, 96, 237.
Spyridaki K, et. al., Structural-Functional Analysis of the Third Transmembrane Domain of the CRF1 Receptor: Role in Activation and Allosteric Antagonism. Journal of Biological Chemistry,2014, 289, 18966.
Matsoukas M-T, et. al. Insights into AT1 Receptor Activation through AngII Binding Studies. Journal of Chemical Information and Modeling,2013, 53, 2798.
Matsoukas M-T, et. al., Ligand Binding Determinants for Angiotensin II Type 1 Receptor from Computer Simulations. Journal of Chemical Information and Modeling,2013,53, 2874.